Performance Profiling Tools for Large-Scale Scientific Applications
Introduction
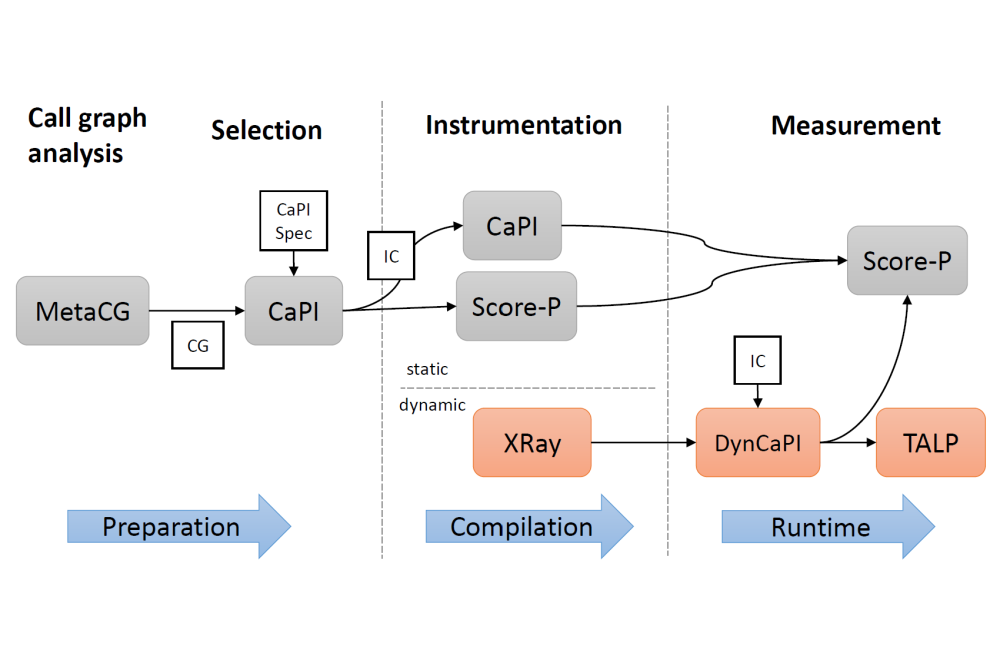
The exaFOAM project (https://exafoam.eu) aims at overcoming the current limitations of Computational Fluid Dynamics (CFD) technology, especially in what concerns the exploitation of massively parallel HPC architectures. This involves the development and validation of a range of algorithmic improvements, across the entire CFD process chain (preprocessing, simulation, I/O, post-processing). The performance assessment of both existing and newly developed code is essential in order to find scalability issues and guide further development. To this end, we are developing specialized performance measurement tooling, aimed at large-scale scientific codes. The use of HPC resources is vital in the assessment of these new tools and their application on OpenFOAM benchmark cases. Within this project period, we extended our previously developed performance profiling tool, called CaPI (https://github.com/tudasc/CaPI), with capabilities for dynamic instrumentation of large-scale scientific applications. This tool enables the creation of performance profiles with minimal measurement overhead.
Methods
We used the Lichtenberg II cluster to evaluate the performance of OpenFOAM benchmarks and the capabilities of our newly developed performance tooling. For dynamic instrumentation of the target code, CaPI now supports the XRay feature of the Clang/LLVM compiler infrastructure. This feature enables the user to adjust the measurement configuration without recompilation by reserving space for measurement calls in the program binary using so-called NO-OP sleds, which can be selectively patched at runtime. We extended XRay with support for shared library instrumentation, thus enabling the instrumentation of OpenFOAM and other scientific applications that rely on such libraries. This new version of CaPI was also extended to be able to interface and direct measurements with the TALP profiling tool. Furthermore, in order to assess the scalability of OpenFOAM and detect potential issues, we performed scaling experiments using instrumented builds of OpenFOAM. To this end, we created empirical performance models of the selected benchmarks using the Extra-P platform.
Results
CaPI was successfully extended with dynamic instrumentation capabilities, greatly improving the profiling workflow applied in exaFOAM. Performance evaluation of the selective dynamic instrumentation approach on Lichtenberg II demonstrated a significant reduction in measurement overhead, while retaining profiling information about performance-critical hotspots. This improvement was leveraged to profile and analyze several OpenFOAM benchmarks with industrial significance. Evaluation of the empirical performance models helped detect a critical scalability bug in OpenFOAM that severely limited its performance on HPC systems. This bug has since been eliminated, leading to a large performance improvement for certain cases.
Discussion
The automation provided by CaPI enabled us to build instrumented configurations of OpenFOAM that were used for the empirical performance modeling of benchmark cases. The improved profiling toolchain enabled us to pinpoint performance and scaling issues faster. Future work includes further extensions to the CaPI profiling infrastructure, such as support for selective tracing with Extrae.