Simulation Structural Plasticity
Introduction
The human brain is a large structure (billions of neurons and orders of magnitude more synapses) that is not understood in its entirety. One possible way of modeling the brain as a whole is assuming that each neuron has its unique need for activity—the change of the individual neurons then drives the change in the whole brain.
These simulations pave the way for modeling memory, learning, and diseases like Alzheimer’s and gliomas. To achieve a meaningful level of meaningful detail, we simulate millions of neurons that exceed the capabilities of current workstations.
Methods
We use the Model of Structural Plasticity and its approximations with variants of the Barnes–Hut algorithm or the Fast Multipole Method.The software is developed by our group, so we can adapt it to our needs when necessary. Additionally, for analyzing the structure of the resulting network, we use the Brain Connectivity Toolbox or an adapted version of it—also developed by our group.
Results
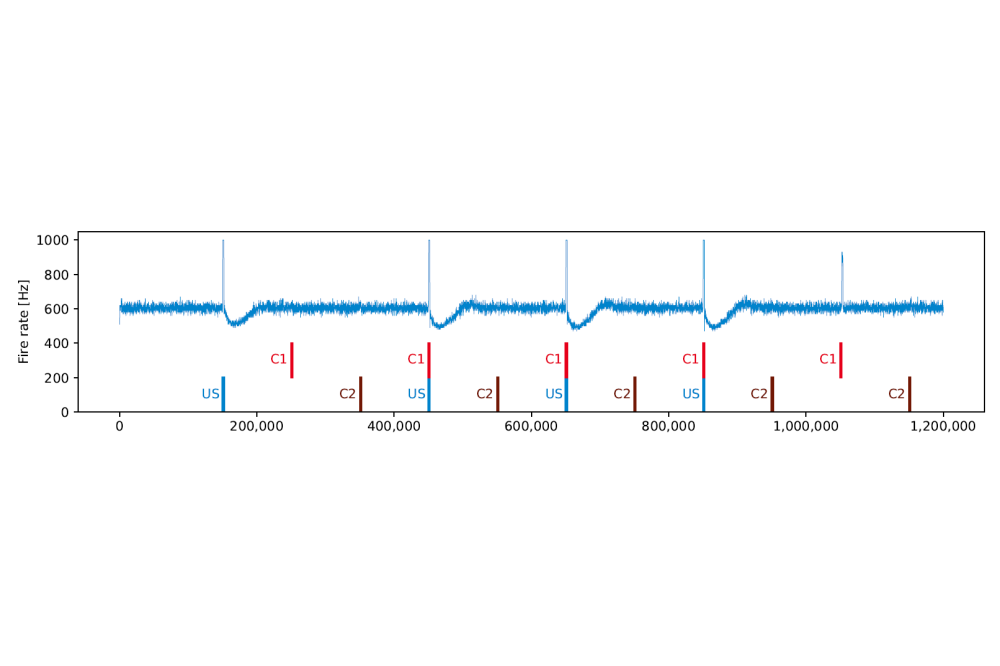
We successfully showed that we can use the Model of Structural plasticity to form silent memory engrams without using a Hebbian learning rule. Furthermore, we could simulate the brain’s input in a more detailed manner than just providing every neuron with a static stimulus. We have shown that we can substitute variant of the Barnes–Hut algorithm with a variant of the Fast Multipole Method without compromising the stability of the stimulation. Furthermore, we succeeded in reducing the communication overhead of the variant of the Barnes–Hut algorithm by approx. 20 %.
Discussion
In this project period, we validated our simulation with biological results, strengthening its stand as a tool to predict and analyze brain behavior. This may lead to further simulations and lastly result in a model for deteriorating brain diseases like Alzheimer’s or various gliomas.
Furthermore, reducing the runtime of our algorithm as well as providing a suitable replacement in the form of the Fast Multipole Method gives users and researchers the freedom to choose according to their needs, as well as reduces the time until their results are ready significantly.