Modeling Soft and Biological Matter Systems
Introduction
The project was majorly dedicated to build cell mechanistic models and study cell mechanics and cell migration. Both models are aimed at capturing both membrane mechanics and cytoskeletal mechanics. First problem is to study the role of cell membrane deformations during immunological synapse. During immunological synapse, the centrosome migrates towards immunological synapse complex. This is achieved by dynein motors walking along the microtubules. We are interested in understanding if the membrane deformability has any role in this processes. Another problem is to study cell migration in complex environments such as tight constrictions or microfluidic type assays.
For immunological synapse problem, cell model will consist of membrane, modelled as elastic triangulated membrane with few thousand vertices, microtubules modelled as semi-flexible filaments and nucleus modelled as stiff triangulated surface. The cell model for second problem will consist of fluidic membrane, again modelled as triangulated surface with around 6000-10000 vertices, and acto-myosin dynamical equations solved on the membrane. In this case, the membrane is fluid which means the topology of connections is not fixed but changing with time, giving rise to fluidic nature of the membrane.
Methods
The primary methodology is particle based simulations. The cell model which is being used for studying immunological synapse problem is implemented in LAMMPS by adding custom fixes. LAMMPS is freely available molecular dynamics package, mainly used for coarse-grained systems. The package is by default written in parallel using MPI scheme. The second cell model which is being used for studying cell migration is written in custom c/c++ code. It is written using openMP parallel scheme.
Results
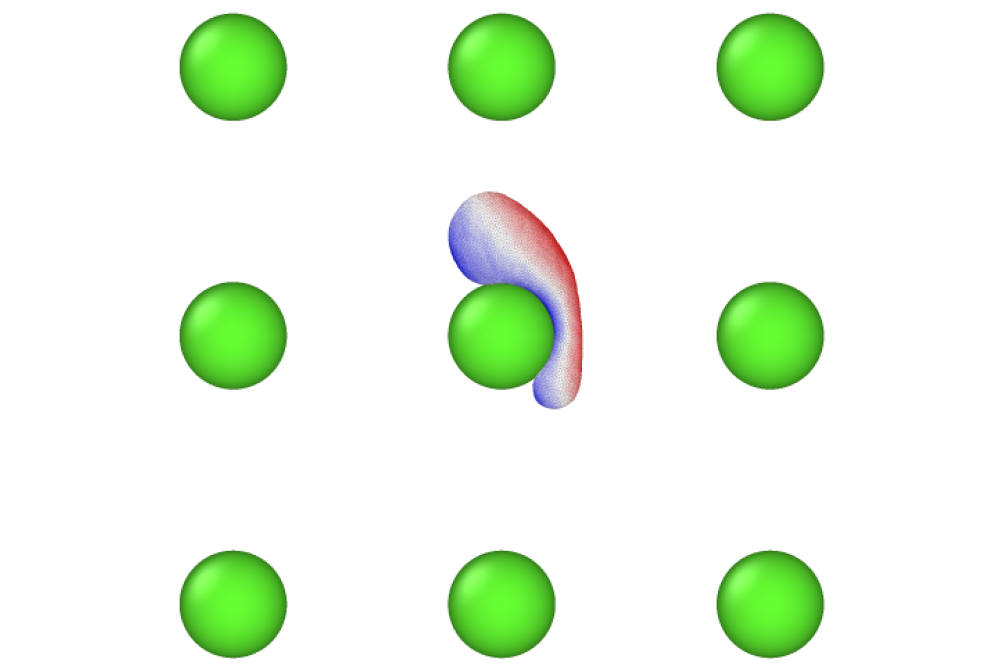
For cell migration problem, the model that accounts for both cell membrane and cytoskeletal mechanics have been built. The model produces typical migration modes that have been observed for keratocyte cells. The migration modes can be classified to amoeboid type migration rather than mesenchymal migration as they don’t produce filopodia kind of structures. These distinct migration modes result from few parameters that model acto-myosin dynamics such as critical angle that limits actin production at the membrane. The membrane elasticity such as bending energy has very weak contribution, but membrane viscosity is a vital parameter. The cell migration speed is observed to be maximum for an optimal critical angle. For immunological synapse problem, the model is successfully implemented in LAMMPS, both in two and three dimensions. I plan to spend new months in producing the results.
Discussion
Computational cell model that is intended to study cell migration has been developed. In the preliminary results, it has been observed that depending on model parameters distinct migration modes can be observed. At the moment, we are constructing a state diagram that accounts for all possible migration modes. Next step is to study the cell migration in inhomogeneous media such as matrix of micro-pillars, constrictions and microfluidic type assays.