Computational Modeling of Biomolecules
Introduction
The rational design and engineering of biomolecules can be support quite often by in silico investigations leveraging powerful compute architectures. These rationale design choices steer experimental verification experiments and support the research focus synthetic biology at TU Darmstadt. In particular, we a) help our experimental colleagues to avoid unnecessary experiments and save costs, but also b) develop new hypotheses on the molecular mechanisms underlying the living cell, which frequently cannot be investigated experimentally, yet. We used the available system to address several, related questions: - ion channel design and modularity - molecular evolution and designability of biotechnological relevant enzymes.
Results
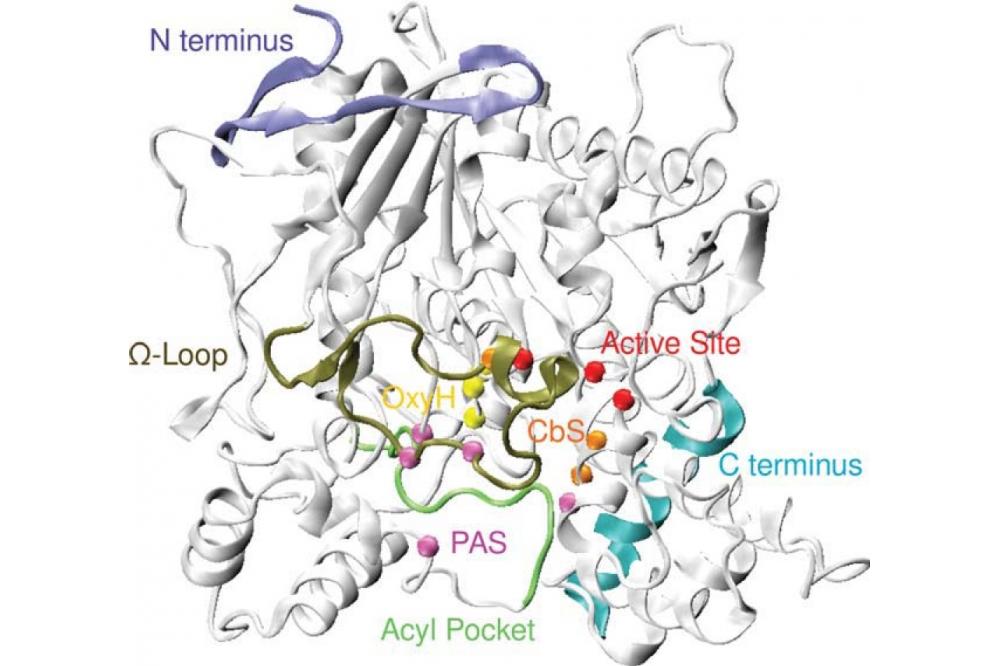
First, we implemented an effi cient, parallelized code to assess co-evolutionary measures, such as the mutual information, efficiently on multi-core architectures, especially GPUs. We developed an algorithm and proved its convergence properties, as well as implemented it, using it for publications and theses [1, 2]. This approach allowed us to understand the molecular evolution of ion channels, e.g. like the viral Kcv channel, in detail. From there, we used reduced molecular models and transferable parametrization of these eff ective theories to scan the „design space“ of various proteins for functional optimization: for ion channels like HCN [3] or for DNA repair proteins like Rad54 [4]. Detailed methodology development [5, 6] allowed us to analyze simulation results under the special requirements of biomolecular engineering processes - as is, e.g., common in our study program „B.Sc./M.Sc. Biomolecular Engineering“ [7, 8]. Eventually leading to new insight on enzymes: e.g. in (Cordes) [9] we showed the close function-evolution relation of the omega-loop in acetylcholinesterase (Fig. 1) - an important drug target and involved in several neurobiological processes.
Outlook
In our work, we tremendously benefit from the available compute resources and the support by the Lichtenberg Cluster and the respective infrastructure of hard-, soft-, and most importantly brainware. We hope to leverage this in the future and collaborate even more on fundamental questions in algorithms and parallelization based upon close collaboration with the groups of Michael Goesele and Andreas Koch, both Deptartment of Computer Science, TU Darmstadt. In particular, on-line reparametrization approaches of the above mentioned coarse-grained models will be the next big issues we want to address, so as to have adaptive codes for large(r) biomolecular complexes.