Learning by Building: Construction of Voltage-Dependent Ion Channels From Modular Components
Introduction
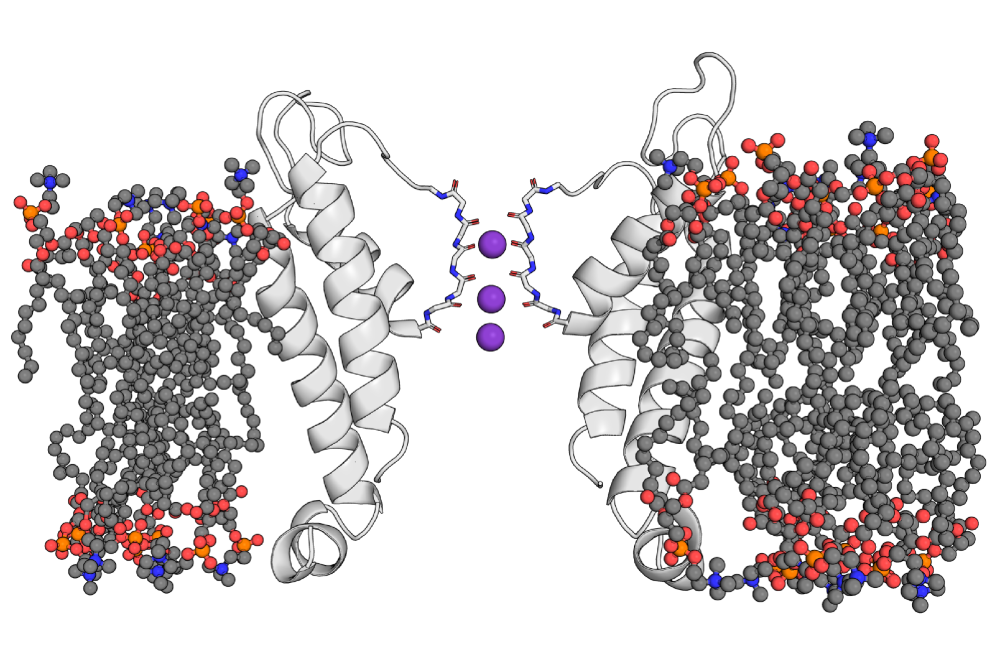
As facilitators of ionic currents across biological membranes, voltage-dependent K+ channels are involved in a broad range of physiological processes, most notably in neuronal signalling. Changes in transmembrane potentials are detected by voltage-sensing domains (VSDs), which open or close the central pore domain following structural rearrangements.
To investigate the basic mechanism of electromechanical coupling between pore, linker and voltage sensor in Kv channels, we will analyse synthetic, chimeric ion channels consisting of the small voltage-insensitive K+ channel Kcv and the voltage sensor domain originating from the phosphatase of the tunicate Ciona intestinalis (Ci-VSD) with computational methods.
Methods
To predict Kcv-Ci-VSD structures, homology modeling approaches and neural network-based methods will be employed.
A combination of coarse-grained elastic network models (ENM) and atomistic molecular dynamics (MD) simulations is employed to extract mechanistic information underlying channel gating. In ENMs, single amino acids are represented as vertices, with covalent and non-bonded interactions between spatially adjacent residues modeled as harmonic springs, whereas movements of single atoms are approximated by classical mechanics in MD simulations. The former retraces global protein dynamics, while the latter resolves motions in atomic resolution. Combined, both methods should allow an extensive analysis of Kcv-Ci-VSD gating with motions occurring on different timescales.
Both structure prediction and MD simulations are computationally costly and can benefit greatly from GPU-acceleration. Therefore, HPC resources are essential to this project.
Results
In the preceeding project period, protein dynamics of the Kcv pore homology structure and of the Ci-VSD were studied separately as reference for the united chimeric structure. Results for ENM-predicted coarse-grained dynamics agree qualitatively with published results for these components. With neural-network based structure prediction methods, chimeric Kcv-Ci-VSD structures were generated. Interestingly, structural differences between these predicted models and older homology models in the N-terminus of the Kcv pore region were observed.
Discussion
Our initial results underline the applicability of ENM-based methods to study coarse-grained dynamics in single structure modules of ion channels. With divergent structures predicted for Kcv using homology-modeling methods and newer neural-network based ones, further scrutiny is required in this regard. To this end, additional methods of structure prediction should be considered.
Outlook
In this project period, we will continue the construction and analysis of Kcv-Ci-VSD homology models. We plan to use recently published software like AlphaFold2, ESM-Fold or RosettaFold together with existing homology-modeled structures of the Kcv pore. Protein dynamics of these constructs will be analysed primarily by ENM-based methods as well as MD simulations. Further insights will be drawn from comparisons with hyperolarization-activated cyclic-nucleotide gated (HCN) ion channels, which are primarily voltage gated, but additionally modulated by cAMP-binding.