Mechanism of TMEM175 Proton Activation
Einleitung
ransmembrane protein 175 (TMEM175) is a cation channel ubiquitously expressed on endosomal and lysosomal membranes. Genetic variations in TMEM175 have been identified as a significant risk factor for neurodegenerative diseases, particularly Parkinson’s disease, establishing it as a promising therapeutic target for small-molecule drug development. Consequently, elucidating the structure-function relationship of TMEM175 is essential to facilitate rational drug design approaches. TMEM175 exhibits pH-dependent ion selectivity: at physiological pH it functions primarily as a K+ channel, whereas acidic conditions in the lysosomal lumen shift selectivity towards H+ conductance. Ion permeation is gated by voltage-dependent mechanisms, enabling pH-controlled regulation of ion flux across the lysosomal membrane. This pH- and voltage-dependent gating mechanism is fundamental to understanding TMEM175’s physiological role and molecular function. Accordingly, we investigated the structural basis underlying this pH-dependent selectivity and gating behavior.
Methoden
To investigate the dynamics of channel opening and closing, we employed molecular dynamics (MD) simulations. MD simulations numerically integrate Newton’s equations of motion using predefined force field parameters, enabling us to observe protein dynamics at high temporal and spatial resolution. However, such simulations are computationally demanding as they require iteratively calculating forces, accelerations, and positions for thousands of atoms (including the surrounding membrane and solvent molecules) across approximately 500 million time steps. This computational cost necessitates substantial processing resources, making GPU acceleration and multi-core computing essential. For these central part of our work, HPC resources provided by the Lichtenberg high performance cluster are essential.
Ergebnisse
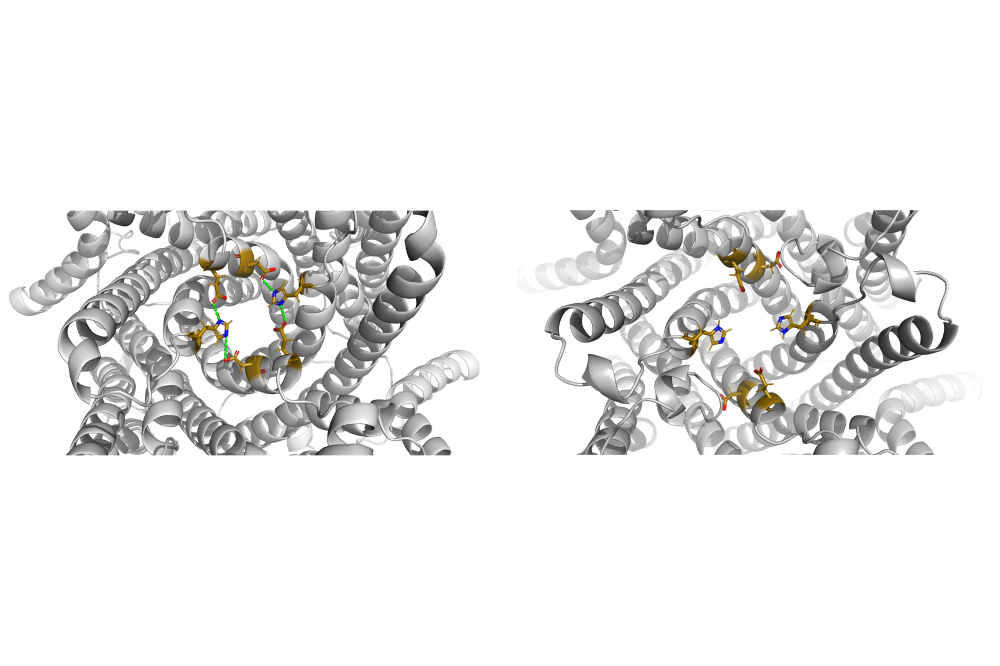
Previous studies identified histidine 57 (H57) as a critical residue governing TMEM175 gating. To elucidate the pH-dependent gating mechanism, we performed MD simulations of the open and closed channel conformations with different protonation states of H57. Our analysis revealed that at lower pH, H57 in the open channel conformation attained critical distances to partner residues for salt bridge formation with significantly higher frequency compared to more basic pH conditions. Conversely, higher pH resulted in reduced salt bridge occupancy. These salt bridges formed by H57 stabilize the open conformation and maintain ion accessibility to the pore. However, the data suggests that H57 is not the sole determinant of channel gating. Additional structural elements likely contribute to the pH-dependent regulation of TMEM175, warranting further investigation.
Diskussion
Although our MD simulations have partially elucidated the gating
mechanism of TMEM175, some questions remain unresolved. In particular, the simulations alone are insufficient to fully explain the observed ion conductance through the channel. Consequently, additional structural investigations are needed to identify the molecular determinants governing pH-dependent channel regulation. Advanced MD simulations employing explicit representation of hydronium ions (H3O+) would enable direct observation of proton and K+ competition, potentially providing crucial mechanistic insights into TMEM175 gating.